Hello again! It’s been a while since my last post and there’s a lot I’d liked to have talked about more if I’d found the time, but with the end of the coding period only a handful of days away, let’s take a look at what I’ve managed to achieve throughout GsoC.
My GSoC project has been divided into three main parts; you can find all the code I’ve been writing (including documentation, examples, and discussion!) by following the links below to the pull request I made for each, and I’ll also give a little summary below that about what’s possbile with each part and the possible future changes and improvements (in addition to general things like more testing and fixing any bugs that pop up). Only one part has reached the stage of merging with MDAnalysis, but the current versions of the other two are at least working and, with a bit more work post-GSoC will hopefully follow soon!
Links to code (pull requests)
-
Add auxiliary – MERGED!
-
Make bundle - in progress
-
Run WHAM - in progress
Add Auxiliary
This part involved adding an ‘auxiliary’ module to MDAnalysis to allow additional timeseries data from a simulation to be read alongside the trajectory. You can read more about it in this post, or to see more about how it’d work in practice, here’s the documentation.

Make bundle
This part (originally planned as a less-general ‘umbrella class’) involved adding a function to let us group together any related simulations and their various ‘metadata’ and ‘auxiliary data’, to make it easier to keep track of and perform analysis across these simulations. You can read more about it in this post.

Run WHAM
The last part involved writing a function to allow WHAM analysis to be performed in MDAnalysis. I didn’t get around to making a post looking at this part in particular, but you can check out the original discussion on github. I mentioned WHAM back in this post; it’s an algorithm we can use to take data from a set of umbrella sampling simulations and calculate the ‘Potential of Mean Force (PMF)’ profile showing the (one-dimensional) energy landscape of the system we’re simulating.
Rather than write a new implementation, I’ve been writing a wrapper for Alan Grossfield’s implementation. Grossfield’s WHAM is run from the command line and uses input files following a particular format. The idea of my wrapper is to allow WHAM to be run in a python script alongside other analysis, remove the need for specific file-formatting, and offer some additional options such as start and end times or changing energy units.

Umbrella Sampling: bringing it all together
The main personal driving force behind my project was to be able to simplify analysis of Umbrella Sampling simulations in MDAnalysis. So how does the work I’ve done pull together to achieve that? Let’s let our final kitties of GSoC demonstrate:
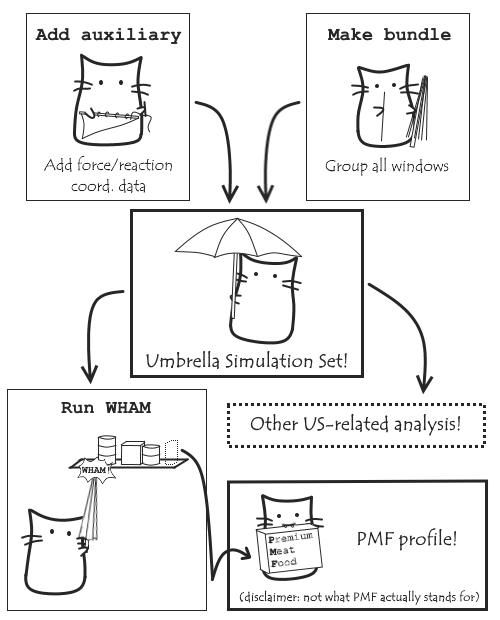
(Can’t remember why we want Umbrella Sampling simulations and PMF profiles? Go back to this post).
Again, there’s still a little work left before this is fully realised within MDAnalysis – but when I find time post-GSoC I’ll definitely be working to get this done!
And finally…
Despite many moments of frustration I’ve enjoyed GsoC: I’ve definitely improved my coding skills and I’ve (mostly) built something I’m likely to actually use for my own research! A huge thanks to my mentors and everyone else at MDAnalysis for help, ideas and discussion along the way.
I had a lot of fun drawing my little cat friends help make this blog interesting, hopefully it’s been fun and informative for you as well! I’d like to keep up posting here (though it’s likely to be even less frequently), so keep an eye out if you’re interested - at the very least, I’ll make a post when ‘make bundle’ and ‘run wham’ are finished.
Thanks for following along GSoC with me and for putting up with all my cats, and (hopefully) see you again sometime!