Hello again! Sorry for the long wait since last post – I’m falling behind my proposed timeline so I’ve been focusing more on trying to catch that up a bit.
I’ve finished work on ‘add_auxiliary’ for now - the general framework and specific case for reading xvg files are basically done, though we’ll see in the coming stages if there are any bugs still to iron out. I’ll probably include in a future post a demonstration of the various features!
Let’s take a look at a revised timeline:
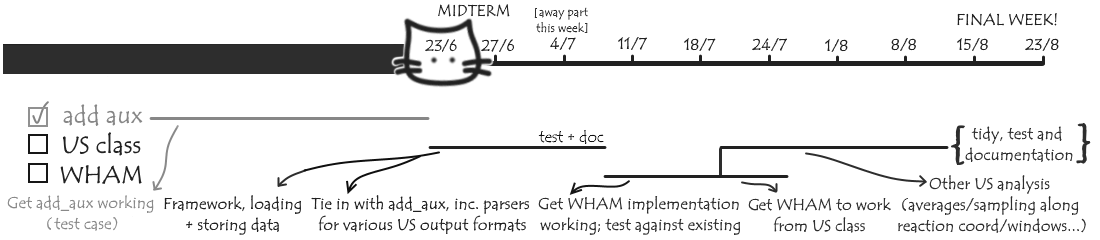
You can see I’m not quite as far along as I was hoping back at the start - building all the add_auxiliary stuff took longer than I expected! I also did some things I originally planned to do later, but in retrospect made more sense to do now. This includes in particular getting the documentation and unit tests nicely done up for AuxReader, and now that I have a good idea how both work, documentation and testing for future parts will hopefully go a lot quicker and smoother! I’m hoping I can stay roughly on track from here - but various bits can be simplified/dropped if need be, and I should still have a nice foundation, which can be further built upon later, by the end of GSoC.
So what next? I’m starting working on part 2 - an ‘Umbrella class’, a framework for storing the trajectories and associated information of a set of Umbrella Sampling simulations. The next post will focus on my plan for this in more detail - but before I leave, something a bit different!
And now for something completely different
All the way back in my first post I mentioned I’m a keen baker. To make consuming sugary snacks even more exciting, I’ve done in the past a couple of ‘edible voting competitions’ within my research group – SBCB, at the University of Oxford – to decide important matters like the fate of New Zealand’s flag design by eating cookies.

I thought this time I’d try something relevant to my GSoC project, so may I present:

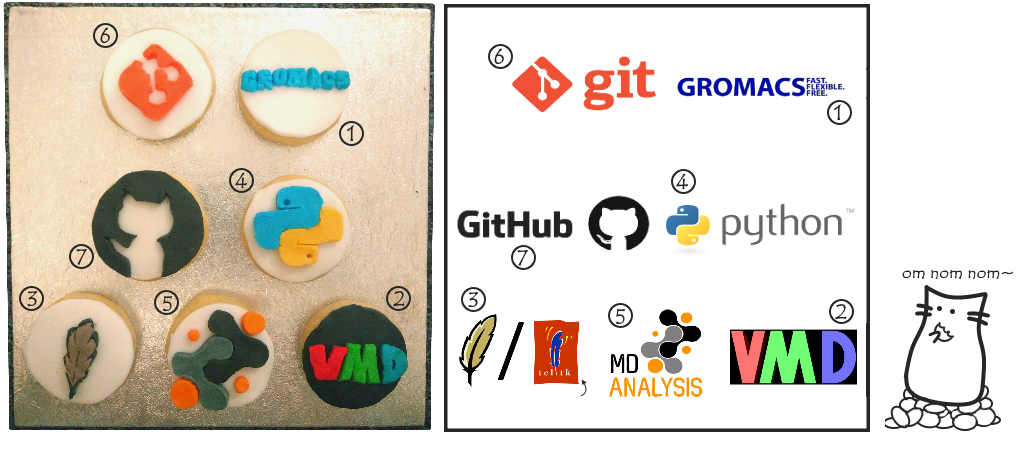
Let’s meet out contestants (the selection of which was heavily biased towards the tools that I use and had ‘logos’ I could reasonably approximate with coloured fondant):
-
GROMACS: Software for performing and analysing Molecular Dynamics (MD) simulations.
-
VMD (Visual Molecular Dynamics): A program for visualising and analysing molecular structures and simulation trajectories.
-
Tcl (and Tk): A programming language; the command-line interface for VMD uses Tcl/Tk, so writing Tcl scripts lets us automate loading and analysing structures/trajectories in VMD.
-
Python: another programming language – as you’re hopefully aware by now, since it’s what I’m using for my GSoC project! In SBCB, often used when writing scripts to automate setup, running and/or analysis of simulations when direct interation with VMD isn’t required.
-
MDAnalysis: A Python library for loading and analysing MD simulations – again, as you hopefully know by now, since it’s what I’m working on for GsoC!
-
Git: A version control system, allowing you to keep track of changes to a set of files in a ‘repository’ (so when you find everything is broken after you made a bunch of changes, you don’t have to spend ages tracking them all down to revert to the working version).
-
Github: A web-based hosting service for Git repositories, allowing sharing of code and collaboration on projects. MDAnalysis is there, I have a page too, and it’s where I’ve been pushing all my proposed changes/additions there (see add_auxilairy here) so other people can check them over!
(There are alternatives for each of these that perform more or less the same functions – but the above are those largely used in SBCB).
We started off with three of each ‘logo’, and the rules were to each time you took a biscuit, take the one you use the least, like the least, or have never even heard of – ideally leaving us with SBCB’s favourite MD tool!
So how did the vote go? (*drumroll*)
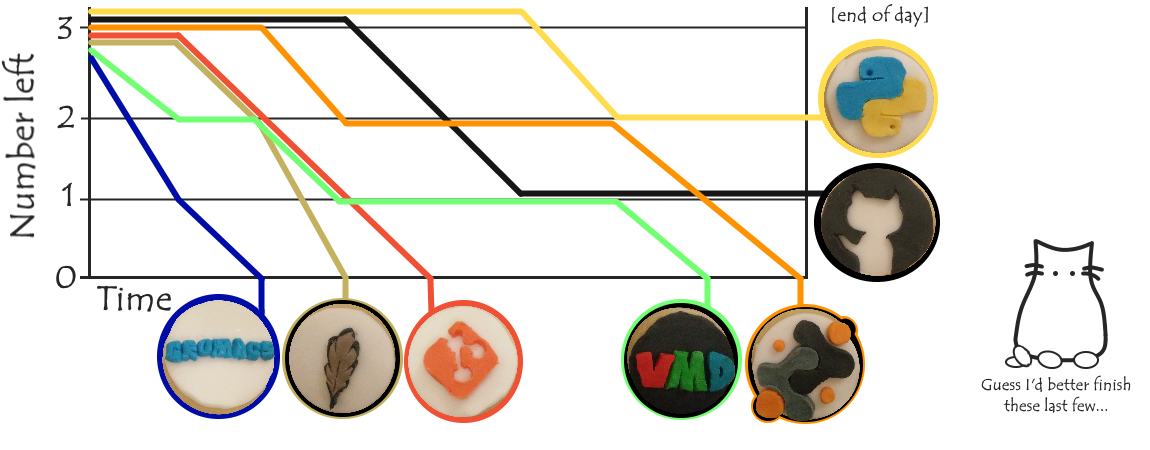
So congratulations Python – you’re the best tool for for MD*, as voted by SBCB!
(*Disclaimer: unfortunately this otherwise highly rigorous ‘scientific study’ was somewhat biased by the participation by several non-MD or non-computational personnel and on a couple of occasions disregard for the rules in favour of the cookie that was closest of most aesthetically pleasing.)
And on that triumphant note, I’ll sign off here, and see you all next post! In the meantime, if you’re disappointed you didn’t get to eat an (unofficial) MDAnalysis cookie, why not go buy an (official, though inedible) MDAnalysis sticker to show off instead?